
2022-03-29 作者:Yingzi Gene
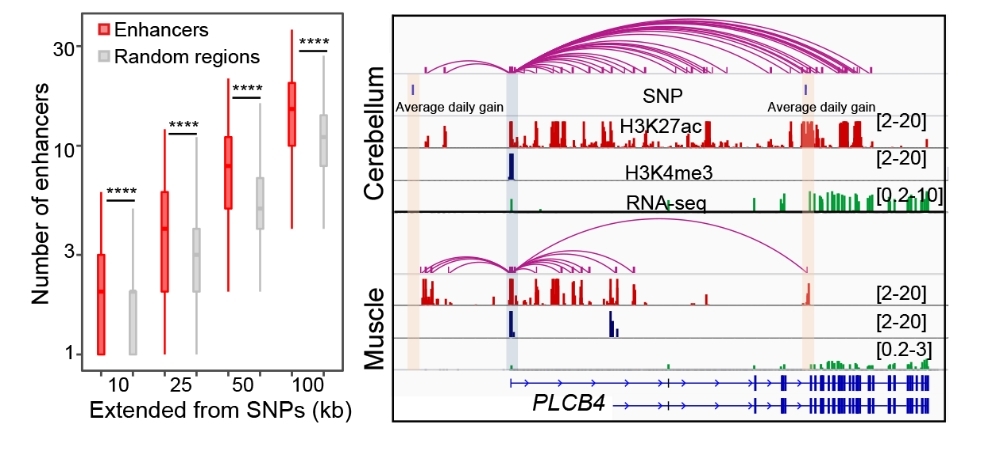
To overcome this bottleneck, Associate Researcher Zhao Yunxia, Dr. Hou Ye, and Ph.D. student Xu Yueyuan, together with their team, have independently established a comprehensive research technology system for studying epigenetic regulation in pig tissues by using several omics technologies such as ChIP-seq, ATAC-seq, RNA-seq, and Hi-C, and obtained 199 sets of epigenetic regulatory data from 12 different types of tissues by taking big lean white pigs, Duroc pigs, fatty Enshi black pigs and Meishan pigs as the research objects. This research has identified over 220,000 regulatory elements and 3,316 novel transcripts in the pig genome, further elucidated the characteristics of super-enhancers and active promoters in the pig genome, and elaborated the tissue specificity of the pig genomic regulatory elements and the impact of their 3D spatial structure on gene expression regulation, which has provided new insights into the functional genomics and trait regulation mechanisms in pigs.

Due to the lag in the exploration of regulatory elements and functional region mutations in the pig genome, the functional loci cannot be adequately distinguished from molecular markers. Consequently, when designing genomic breeding chips, the information related to functional regions and loci cannot be fully utilized, thereby influencing the improvement of the genomic breeding efficiency. This study has integrated regulatory elements, 3D genome, and genome-wide association analysis (GWAS) information to elucidate the distribution characteristics of GWAS loci for important pig traits around regulatory elements, identified candidate causal mutations for some important traits, and also identified a batch of potentially functional SNPs by integrating genomic mutations, epigenetic modifications, 3D genomes and gene expression variations among different breeds. The aforementioned achievements have provided a wealth of data and information for the innovation and improvement of genomic breeding technologies, and opened up new avenues to accelerate genetic enhancements of economic traits in pigs.
[Abstract]
Although major advances in genomics have initiated an exciting new era of research, a lack of information regarding cis-regulatory elements has limited the genetic improvement or manipulation of pigs as a meat source and biomedical model. Here, we systematically characterize cis-regulatory elements and their functions in 12 diverse tissues from four pig breeds by adopting similar strategies as the ENCODE and Roadmap Epigenomics projects, which include RNA-seq, ATAC-seq, and ChIP-seq. In total, we generate 199 datasets and identify more than 220,000 cis-regulatory elements in the pig genome. Surprisingly, we find higher conservation of cis-regulatory elements between human and pig genomes than those between human and mouse genomes. Furthermore, the differences of topologically associating domains between the pig and human genomes are associated with morphological evolution of the head and face. Beyond generating a major new benchmark resource for pig epigenetics, our study provides basic comparative epigenetic data relevant to using pigs as models in human biomedical research.
Paper Link:https://www.nature.com/articles/s41467-021-22448-x